Brain development is a complex and precisely regulated process involving key steps such as proliferation and differentiation of neural stem cells. Developmental abnormalities can lead to severe neurodevelopmental disorders, such as microcephaly, bringing heavy burdens to patients, families, and society. During brain development, neural stem cell proliferation requires active ribosome biogenesis to meet the demand for massive protein synthesis. Ribosome biogenesis is a highly coordinated and energy-consuming process involving the coordinated participation of a series of ribosomal RNA (rRNA) processing factors and ribosome assembly molecules. Existing studies have shown that ribosome biogenesis defects are associated with brain developmental defects such as microcephaly, while enhanced ribosome biogenesis is closely related to cancer occurrence. Therefore, precise regulation of ribosome biogenesis is crucial for both normal development and cancer prevention. However, the RNA epigenetic regulatory mechanisms of ribosome biogenesis are currently unclear.

Paper Link:
https://www.science.org/doi/10.1126/sciadv.adq9643
On June 27, the team led by Associate Professor Zhou Tao from the Faculty of Life and Health Sciences at Shenzhen University of Advanced Technology, together with Professor Shen Bin’s team from Nanjing Medical University, published a paper inScience Advances titled “VIRMA-mediated m6A modification regulates forebrain formation through modulating ribosome biogenesis.”The study reveals for the first time how VIRMA, the core scaffold protein of the m6A methyltransferase complex, influences brain development by regulating ribosome biogenesis.
N6-methyladenosine (m6A) modification, as a widely existing post-transcriptional regulatory mechanism in eukaryotes, has dynamic reversible characteristics jointly regulated by the methyltransferase complex (also known as m6A "writer", including METTL3, METTL14, WTAP, and VIRMA, etc.) and demethylases (also known as m6A "eraser"), and affects RNA metabolism and function through m6A recognition proteins (also known as m6A "reader"). In 2011, Professor Chuan He's team from the University of Chicago discovered the first demethylase FTO, revealing the dynamic reversible nature of m6A and sparking a research boom in RNA epigenetics. Although there are domestic and international studies on m6A modification and embryonic brain development, for example, conditional knockout of the catalytic components METTL3 or METTL14 of the methyltransferase complex can lead to brain developmental defects, however, these studies have significant differences in molecular mechanisms, and the specific mechanisms need to be unified.
As the largest and evolutionarily newest component in the methyltransferase complex, VIRMA has unique structural and functional characteristics. Structural studies show that VIRMA presents a characteristic "horse-shaped" conformation and together with WTAP forms the structural core of the complex. Particularly noteworthy is that three of the four RNA binding sites in the methyltransferase complex are located on VIRMA, making it play a key role in substrate recognition. This study, for the first time, takes VIRMA as the entry point, by constructing conditional gene knockout mice and neural stem cell models, combined with various biochemical analysis methods and multi-omics sequencing (m6A-seq, RNA-seq, Proteomic, etc.) technologies, discovered:
1. Core scaffold function: VIRMA, as the core scaffold protein of the m6A methyltransferase complex, its absence leads to instability of the entire complex, reducing m6A modification levels on mRNA by 60%, disrupting mRNA metabolic homeostasis (Figure 1);
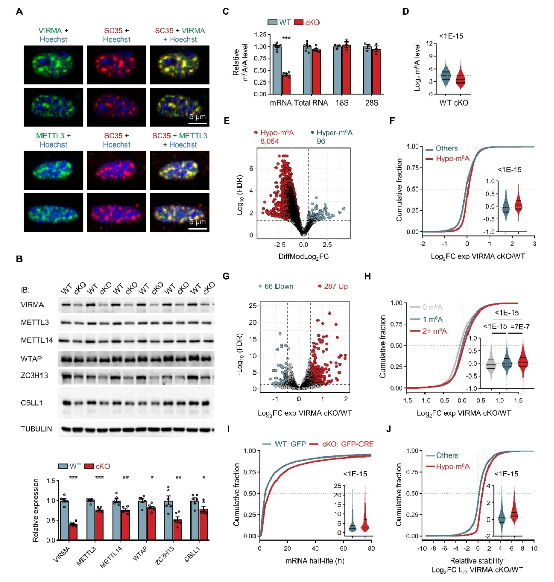
Figure 1. VIRMA Absence Leads to mRNA's
m6A Modification Levels Decrease and Disrupt mRNA Metabolic Homeostasis
2. Dysregulated ribosome biogenesis: Reduced m6A modification prolongs the half-life of key ribosome biogenesis factors (e.g., RPP38, NCL), causing abnormal mRNA accumulation and excessive protein expression, which paradoxically leads to defective ribosome biogenesis and markedly suppressed protein translation (Figure 2).
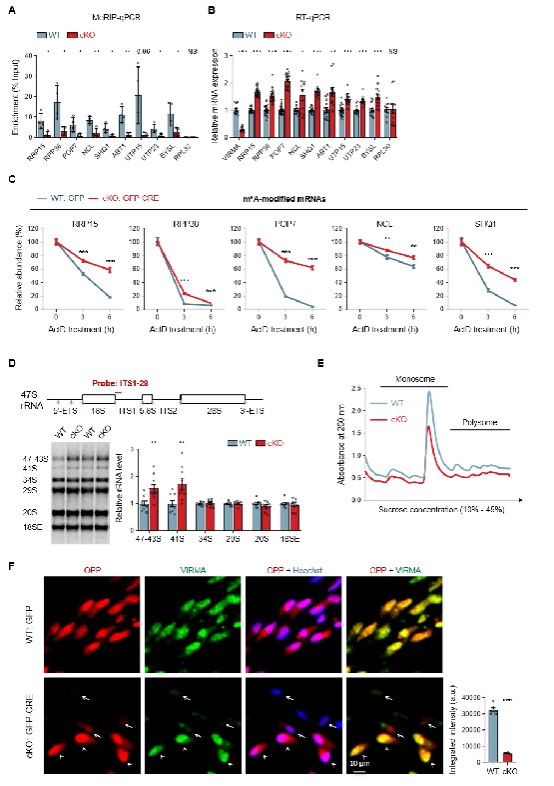
Figure 2. VIRMA Absence Triggers Ribosome Biogenesis Defects and Global Protein Translation Inhibition by Prolonging mRNA Half-Life of Key Ribosome Biogenesis Factors
3. Impaired forebrain development: Ribosome biogenesis defects trigger p53-dependent nucleolar stress response, inhibiting p53 protein degradation, leading to excessive accumulation and activation of p53 in cells, causing cell cycle arrest and apoptosis, resulting in abnormal brain structure (Figure 3).
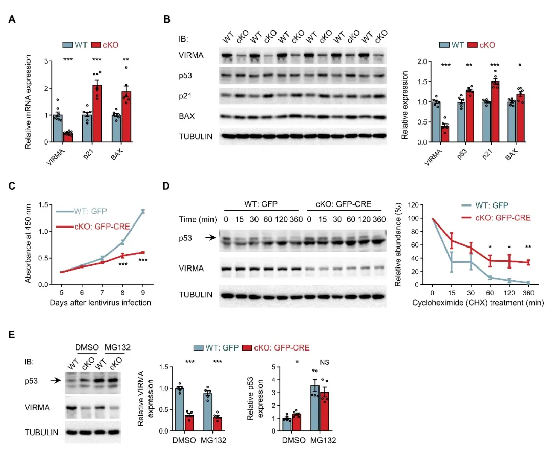
Figure 3. VIRMA Absence Through p53-Dependent Nucleolar Stress Response
Triggers Cell Cycle Arrest and Apoptosis
In summary, this study elucidates the molecular mechanism of the VIRMA-m6A-ribosome axis regulating brain development (Figure 4), providing a new perspective for understanding neurodevelopmental diseases such as microcephaly. In addition, this study preliminarily found that VIRMA also regulates ribosome synthesis in breast cancer cells (MCF7) and cervical cancer cells (HeLa), suggesting its value as a potential therapeutic target.
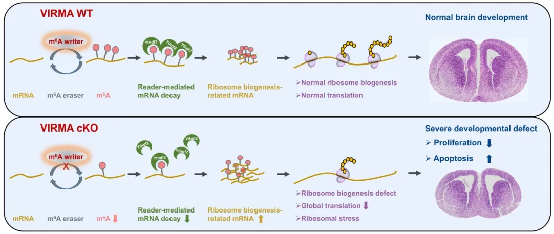
Figure 4. VIRMA-Mediated m6A Modification Regulates Ribosome
Schematic of the molecular mechanism by which the ribosome biogenesis pathway participates in brain development
Associate Professor Zhou Tao and Assistant Professor Wu Min from Shenzhen University of Advanced Technology, together with Professor Shen Bin from Nanjing Medical University, are co-corresponding authors.Assistant Professor Wu Min, PhD student Wu Xiaoli (now postdoctoral fellow at Shenzhen Institute of Advanced Technology), and PhD student Sun Haifeng from Nanjing Medical University (now postdoctoral fellow at Yale University) are co-first authors. This research was completed with funding from the National Key R&D Program of the Ministry of Science and Technology, the National Natural Science Foundation of China, the Guangdong Basic and Applied Basic Research Foundation, and the Shenzhen Science and Technology Innovation Plan.
Postdoctoral Positions Available (Long-term Recruitment)
Our lab has long-term open postdoctoral positions on RNA modifications in the nervous system. Interested candidates in biology/medicine/bioinformatics/neuroscience, please send CV tozhoutao@suat-sz.edu.cn (Zhou Tao).